Section: New Results
Computational Physiology
Computational modeling of radiofrequency ablation for the planning and guidance of abdominal tumor treatment
Participants : Chloé Audigier [Correspondent] , Hervé Delingette, Tommaso Mansi [Siemens] , Nicholas Ayache.
This PhD work was carried out between the Asclepios research group, Inria Sophia Antipolis, France and Medical Imaging Technologies, Healthcare Technology Center, Siemens Medical Solutions USA, Princeton, NJ.
Radiofrequency Ablation Modeling, Patient-Specific Simulation, Lattice Boltzmann Method, Computer Model, Computational Fluid Dynamics, Heat Transfer, Cellular Necrosis, Parameter Estimation, Therapy Planning, Liver, Pre-clinical Study, Medical Imaging
Radio Frequency Ablation (RFA) is a minimally invasive therapy suited for liver tumor ablation. However, a patient-specific predictive tool is needed to plan and guide the treatment.
We developed a computational framework for patient-specific planning of RFA, which includes the following contributions:
-
A detailed computational model of the biophysical mechanisms (heat transfer, cellular necrosis, hepatic blood flow) involved in RFA of abdominal tumors based on patient images.
-
A new implementation of the bio-heat equations coupled with a cellular necrosis model using the Lattice Boltzmann Method (LBM) on Graphics Processing Units (GPU), which allows near real-time computation.
-
A Computational Fluid Dynamics (CFD) and porous media solver using LBM algorithm to compute the patient-specific blood flow in the hepatic circulatory system and the blood flow distribution inside the parenchyma.
-
A complete patient-specific geometry including hepatic venous and arterial circulation system.
-
The automatic estimation of the main parameters of the model. Two personalization strategies tested and evaluated on clinical and pre-clinical data.
-
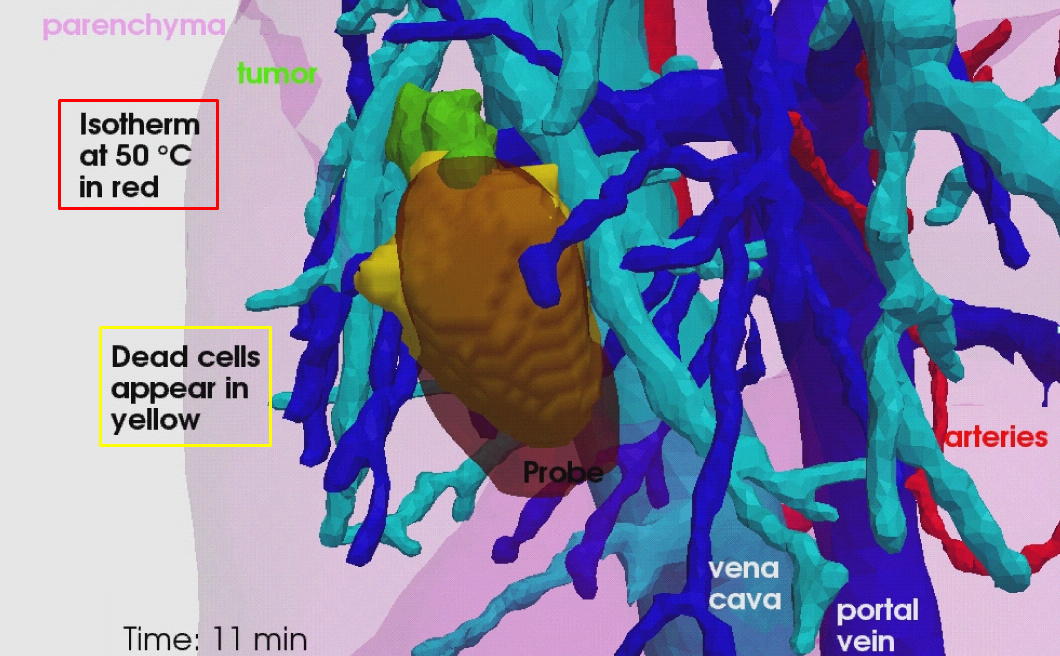
The evaluation of the proposed model on a clinical dataset of ten patients (see Fig. 12 ).
-
The evaluation on a preclinical dataset of five swines from a comprehensive experimental set-up specially designed for RFA model validation.
The proposed RFA model and its evaluation on clinical data are presented in [10] , and the evaluation of the RFA model on pre-clinical data is presented in [25] . The proposed model, its personalisation and its evalutation against clinical and preclinical data are presented in Chloé Audigier's PhD thesis [1] .
Learning Cardiac Ablation Targets from Image Data and Simulation
Participants : Rocio Cabrera Lozoya [Correspondent] , Maxime Sermesant, Nicholas Ayache.
This work was supported by the ERC Advanced Grant MedYMA 2011-291080 (on Biophysical Modeling and Analysis of Dynamic Medical Images).
Cardiac electrophysiology modeling, Intracardiac electrogram modeling, Machine learning, Radiofrequency ablation planning, electroanatomical mapping, local abnormal ventricular activities (LAVA)
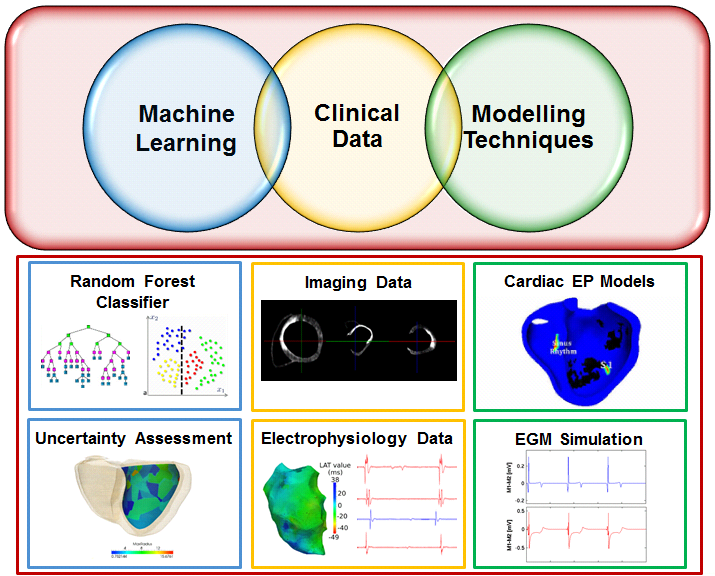
Ventricular radiofrequency ablation can have a critical impact on preventing sudden cardiac arrest but it is challenging due to a highly complex arrhythmogenic substrate. We used advanced delayed enhanced-MR image characteristics in a machine learning framework to predict the presence of local abnormal ventricular activities (LAVA). Furthermore, we enriched these predictions through MR image-based patient-specific electrophysiology simulations and the modeling of normal and LAVA-like intracardiac electrograms using the dipole approach and their incorporation in the learning framework (see Fig. 13 ). Confidence maps can then be generated and analyzed prior to RFA to guide the intervention.
Biophysical Modeling and Simulation of Longitudinal Brain MRIs with Atrophy in Alzheimer's Disease
Participants : Bishesh Khanal [Correspondent] , Nicholas Ayache, Xavier Pennec.
This work has been partly supported by the European Research Council through the ERC Advanced Grant MedYMA (on Biophysical Modeling and Analysis of Dynamic Medical Images).
Alzheimer's Disease (AD), modeling brain deformation, biophysical model, simulation
-
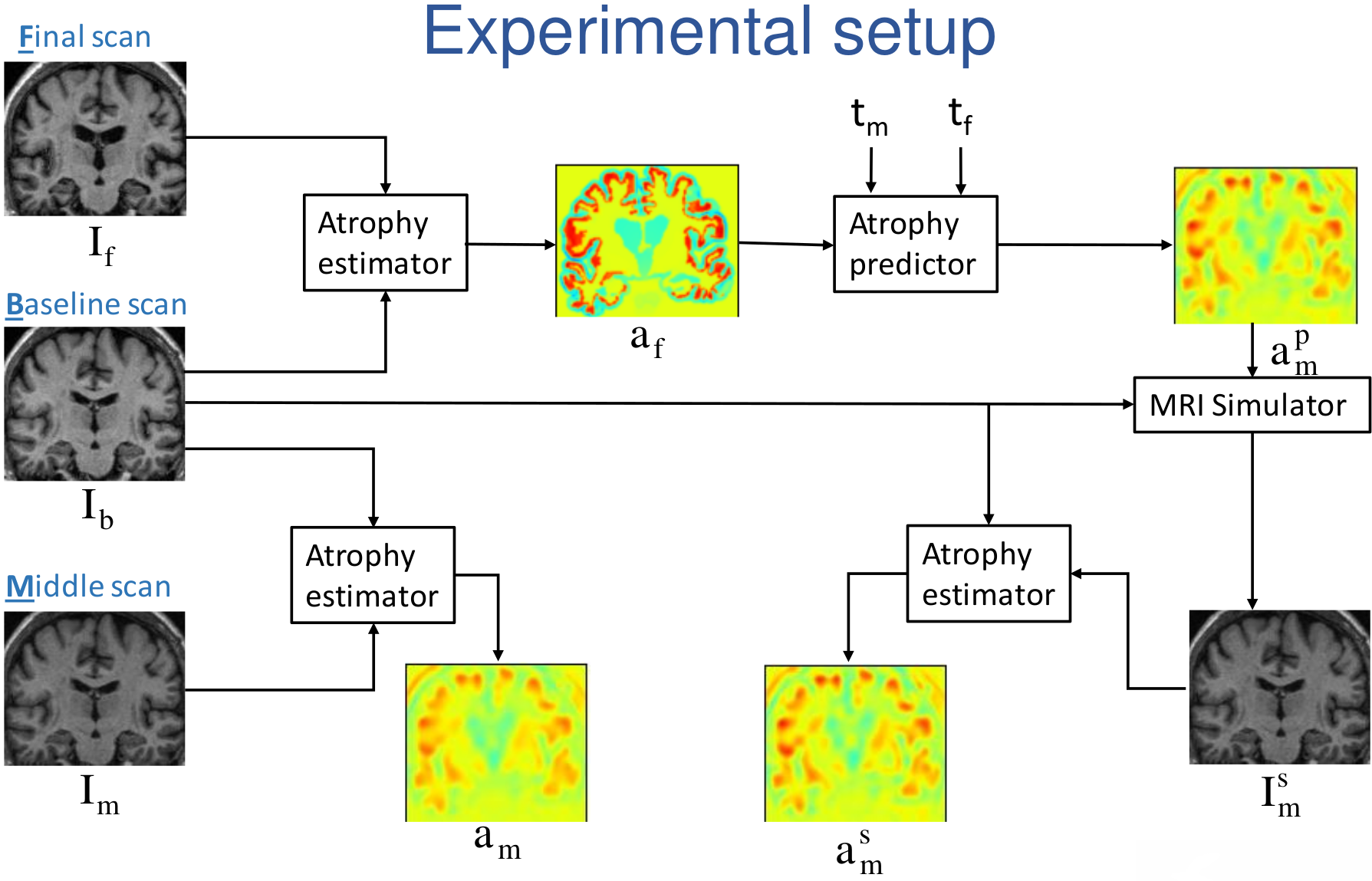
We developed a framework to generate patient specific multiple time-point images based on our biophysical model of brain deformation due to atrophy in Alzheimer's Disease (AD)[34] . From two time-point brain MRIs of a patient, we used the framework to simulate a new time-point brain MRI with the personalized atrophy for the patient (see Fig. 14 ).
-
The framework can be used to evaluate methods that study the temporal relationships, ordering and co-evolution of atrophy in different structures of the brain.
|
Brain Tumor Growth Personalization and Segmentation Uncertainty
Participants : Matthieu Lê [Correspondent] , Hervé Delingette, Jan Unkelbach, Nicholas Ayache.
This work is carried out between Asclepios research group, Inria Sophia Antipolis, France and the Department of Radiation Oncology of the Massachusetts General Hospital, Boston, USA. It is supported by the ERC Advanced Grant MedYMA 2011-291080 (on Biophysical Modeling and Analysis of Dynamic Medical Images).
Tumor growth, radiotherapy, modeling, personalization, segmentation, uncertainty, Bayesian
-
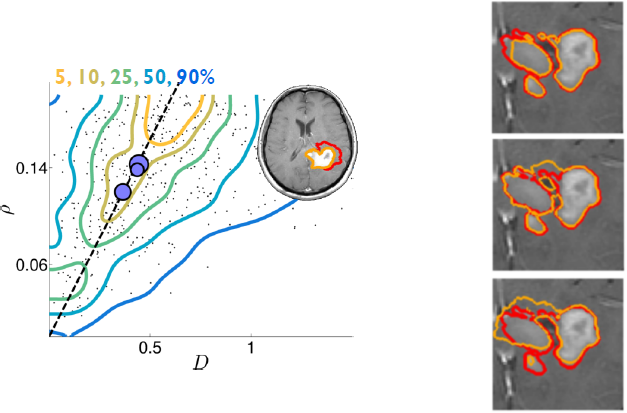
We developed a method for the Bayesian personalization of a brain tumor growth model based on clinical MRIs [37] (see Fig. 15 Left).
-
We proposed an algorithm for the sampling of several plausible segmentations, based on a single clinical segmentation (see Fig. 15 Right). This allows the uncertainty quantification of the radiotherapy plan based on several sample clinical target volumes [38] . This paper received the Young Scientist Award at the 2015 MICCAI conference in Munich, Germany.
|
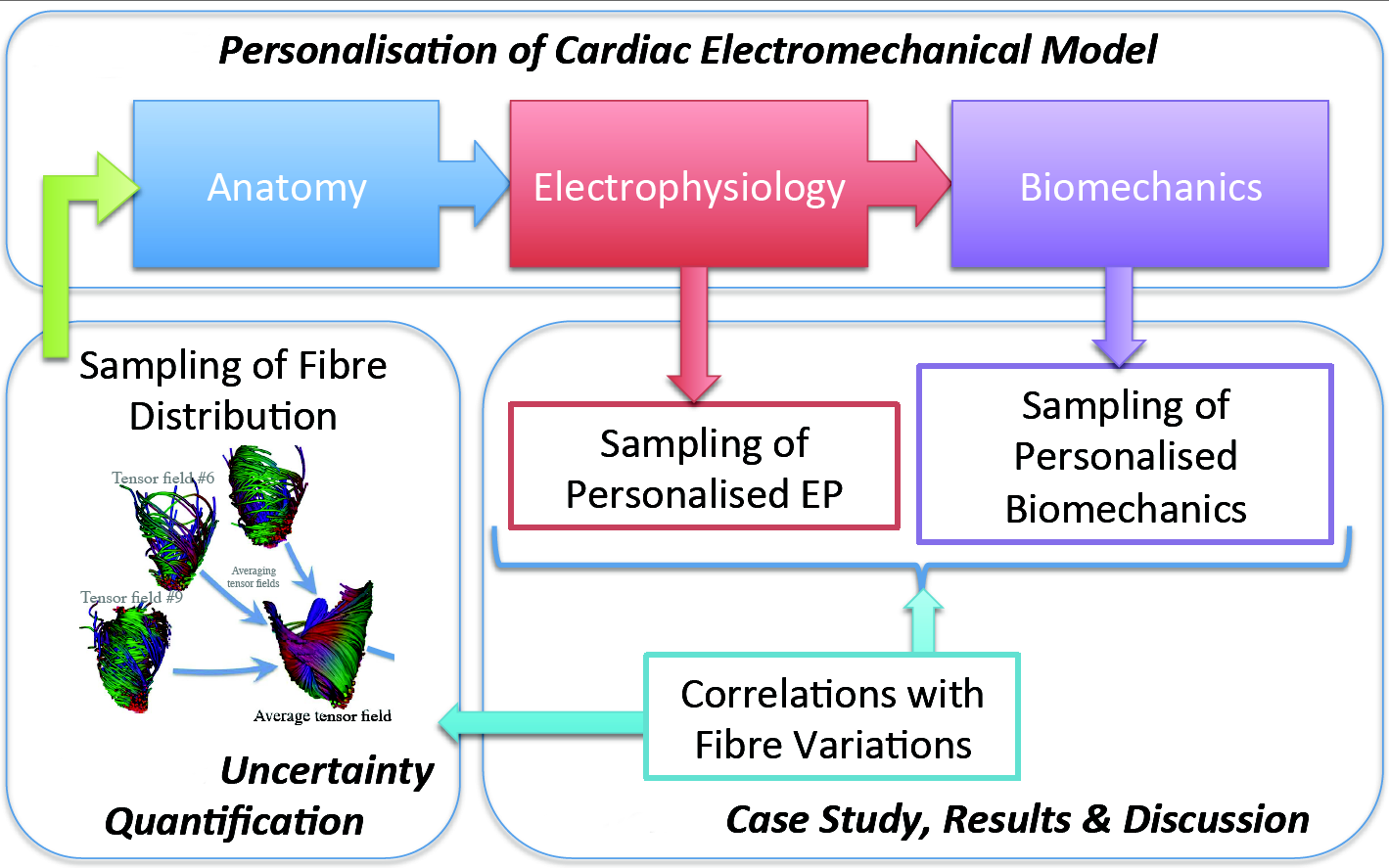
Uncertainty quantification in personalised Cardiac models. Application to myocardial fiber uncertainty.
Participants : Roch-Philippe Molléro [Correspondent] , Dominik Neumann [Siemens] , Marc-Michel Rohé, Hervé Delingette, Maxime Sermesant, Xavier Pennec, Nicholas Ayache, Tommaso Mansi [Siemens] .
This work was partly supported by the FP7 European project MD-Paedigree and was done in collaboration with Siemens Corporate Technology, Erlangen, Germany and Siemens Corporate Research, Princeton, New Jersey.
Heart Modeling - Myocardial Fibers - Biophysical Simulation - Uncertainty Quantification
Computational models of the heart are of increasing interest for clinical applications due to their discriminative and predictive power. However, the personalisation step to go from a generic model to a patient-specific one is still challenging. In particular, it is still difficult to quantify the uncertainty on the estimated parameters and predicted values.
We developped a pipeline (see Fig. 16 ) to evaluate the impact of myocardial fibre uncertainty on the personalisation of an electromechanical model of the heart from ECG and medical images:
-
We studied how to estimate the variability of the fibre architecture among a given population (from a myocardial fiber atlas).
-
Then, we showed the variability of the personalised simulations, in electrophysiology (EP) and in biomechanics, with respect to the principal variations of the fibres.
-
Finally, we discussed how the variations in this population of fibres impact the parameters of the personalised simulations.
This work led to a paper at FIMH 2015 conference in Maastricht, The Netherlands [45] .
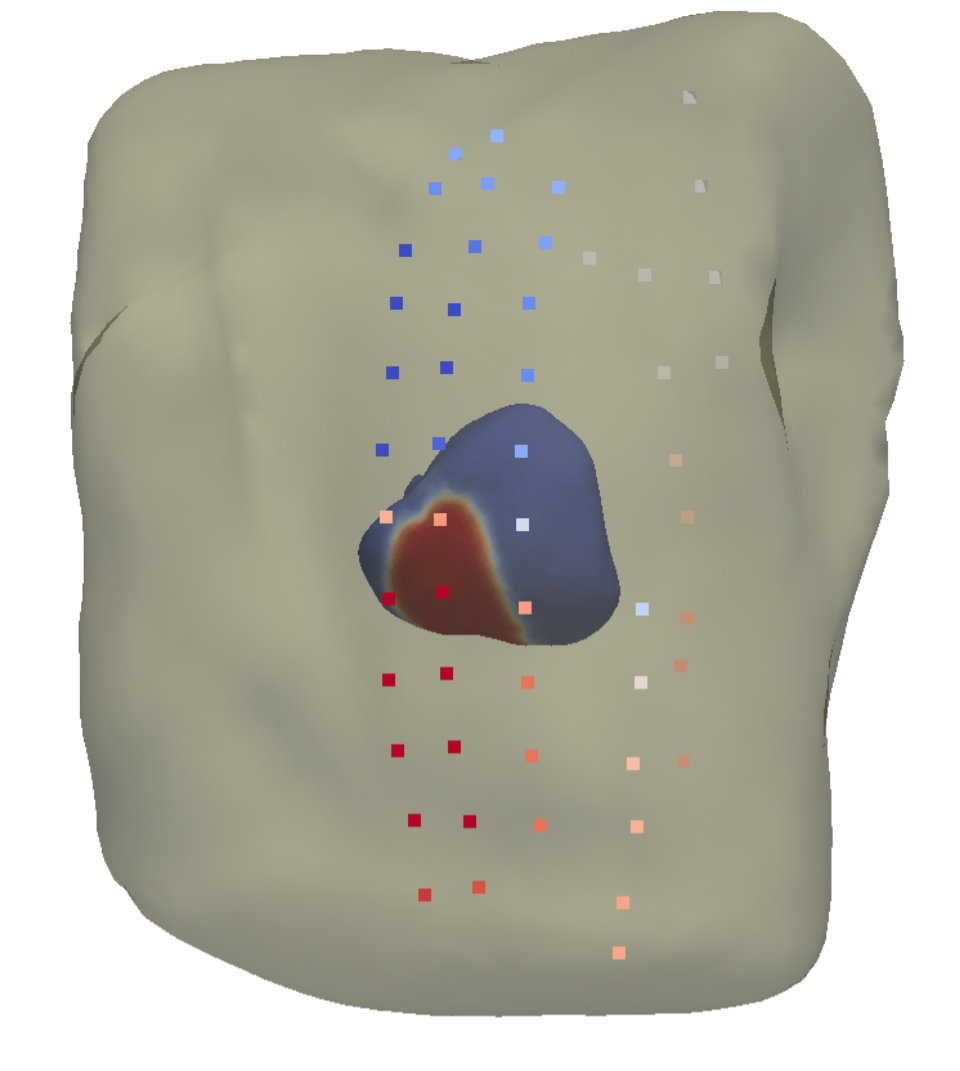
Non-invasive personalisation of the electrical heart model
Participants : Sophie Giffard-Roisin [Correspondent] , Maxime Sermesant, Nicholas Ayache, Hervé Delingette.
This work has been supported by the European Project FP7 under grant agreement VP2HF (no 611823) and the ERC Advanced Grant MedYMA (on Biophysical Modeling and Analysis of Dynamic Medical Images).
Cardiac Modelling, Personalised Simulation, Electrical Simulation
Non-invasive cardiac electrical data has been acquired at St Thomas' Hospital, London. It consists in Body Surface Potential Mapping (BSPM), which are recordings of the electrical potential on several locations on the surface of the torso (see Fig. 17 ). From BSPMs and MRI data of the heart, we aim at personalizing the electrical propagation model of the heart previously developed within the Asclepios team.